Article
 YiRang Shin (left) and Pengfei Song in the Song Lab at the Beckman Institute for Advanced Science and Technology. Credit: Elizabeth Bello, Beckman Institute Communications Office.
Researchers at the Beckman Institute for Advanced Science and Technology developed a new technique to make ultrasound localization microscopy, an emerging diagnostic tool used for high-resolution microvascular imaging, more practical for clinical settings. Their method uses deep learning to advance in the post-processing pipeline of ULM.
YiRang Shin (left) and Pengfei Song in the Song Lab at the Beckman Institute for Advanced Science and Technology. Credit: Elizabeth Bello, Beckman Institute Communications Office.
Researchers at the Beckman Institute for Advanced Science and Technology developed a new technique to make ultrasound localization microscopy, an emerging diagnostic tool used for high-resolution microvascular imaging, more practical for clinical settings. Their method uses deep learning to advance in the post-processing pipeline of ULM.
Their technique, called LOcalization with Context Awareness Ultrasound Localization microscopy, or LOCA-ULM, appears in the journal Nature Communications.
“I’m really excited about making ULM faster and better so that more people will be able to use this technology. I think deep learning-based computational imaging tools will continue to play a major role in pushing the spatial and temporal resolution limits of ULM,” said first author YiRang Shin, a graduate student in the Department of Electrical and Computer Engineering at the University of Illinois Urbana-Champaign.
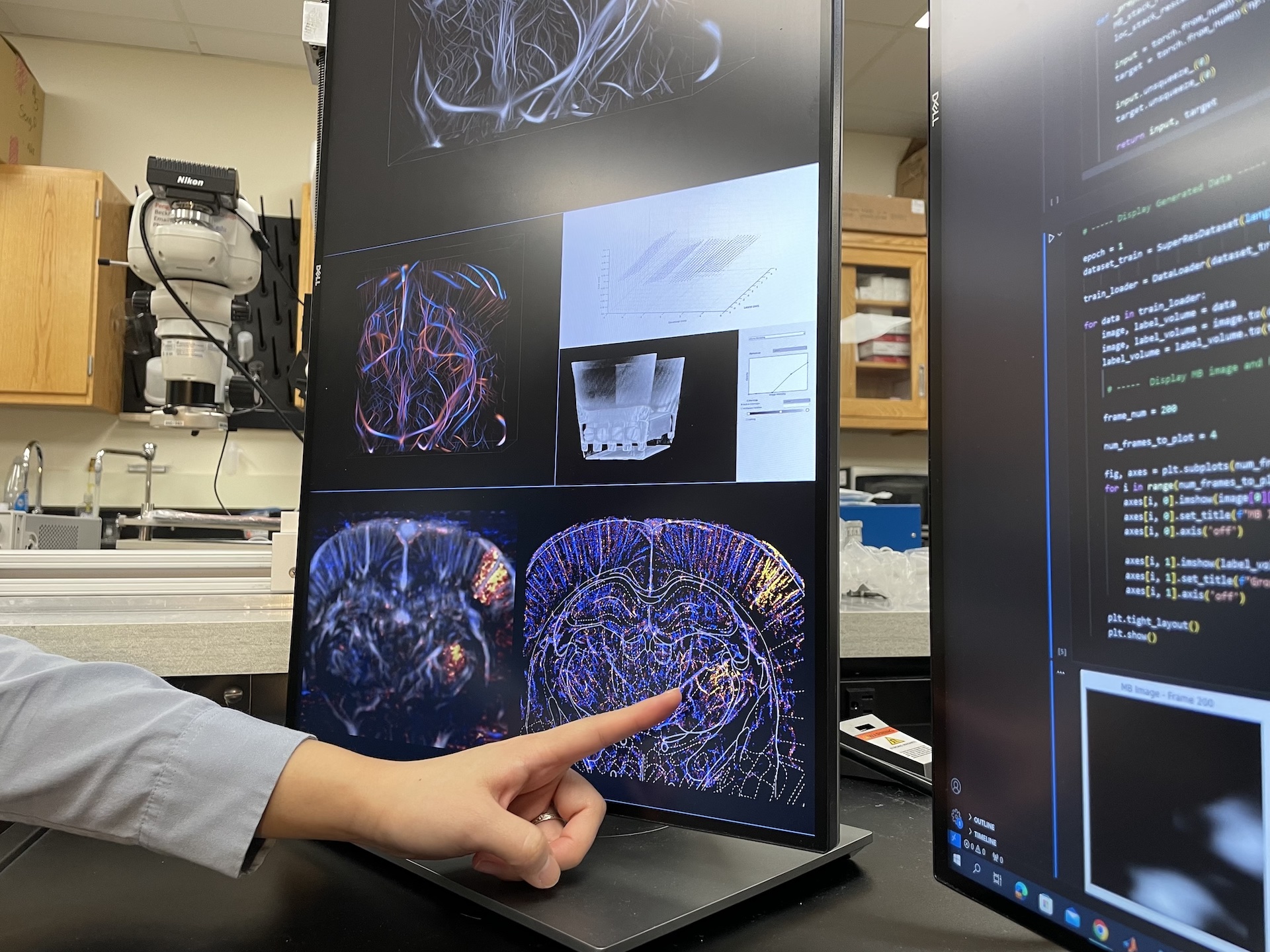 Looking at an ultrasound image, Shin points to a region of the brain showing strong microbubble activity from a nerve stimulation test. Credit: Elizabeth Bello, Beckman Communications Office.
Ultrasound localization microscopy works by injecting microbubbles into blood vessels, where they act as contrast agents. Microbubbles are FDA-approved for clinical use. Ultrasound waves can penetrate deep tissues in the body to pinpoint the location
of these microbubbles — each only several microns in size — as they travel through the bloodstream. Researchers use microbubbles to track blood flow speed and create spatial images of blood vessels at the microscale.
Looking at an ultrasound image, Shin points to a region of the brain showing strong microbubble activity from a nerve stimulation test. Credit: Elizabeth Bello, Beckman Communications Office.
Ultrasound localization microscopy works by injecting microbubbles into blood vessels, where they act as contrast agents. Microbubbles are FDA-approved for clinical use. Ultrasound waves can penetrate deep tissues in the body to pinpoint the location
of these microbubbles — each only several microns in size — as they travel through the bloodstream. Researchers use microbubbles to track blood flow speed and create spatial images of blood vessels at the microscale.
The current imaging speed of ULM has limited its practical application as a diagnostic tool in the medical field and a research tool in basic science. Increasing imaging speed requires a higher concentration of microbubbles in the bloodstream, which makes the post-processing much more difficult, Shin said.
The researchers’ new method demonstrates higher imaging performance and processing speed, increased sensitivity for functional ULM and overall superior in vivo imaging. It also demonstrates improved computational and microbubble localization performance and is adaptable to different microbubble concentrations.
“It really beats conventional microbubble localization methods; this is the way to go,” said Pengfei Song, a Beckman researcher and the Y.T. Lo Faculty Fellow and assistant professor of electrical and computer engineering at Illinois.
To make microbubble localization faster, more accurate and more efficient, the researchers developed a simulation model based on a generative adversarial network called GAN. This simulation creates realistic microbubble signals to train the deep context-aware neural network DECODE.
Editor’s note:
The paper titled “Context-aware deep learning enables high-efficiency localization of high concentration microbubbles for super-resolution ultrasound localization microscopy” can be accessed at: https://doi.org/10.1038/s41467-024-47154-2
Pengfei Song is also an assistant professor of bioengineering and biomedical and translational sciences and an affiliate of the Carl R. Woese Institute for Genomic Biology and the Cancer Center at Illinois.
Additional coauthors include Matthew R. Lowerison, Yike Wang, Xi Chen, Qi You, Zhijie Dong and Mark A. Anastasio. For full author information, please consult the publication.
Media contact: Jenna Kurtzweil, kurtzwe2@illinois.edu
Beckman Institute for Advanced Science and Technology