Article
A new computational mass spectrometry imaging method enables researchers to achieve high mass resolution and high spatial resolution for biological samples while providing data sets exponentially faster.
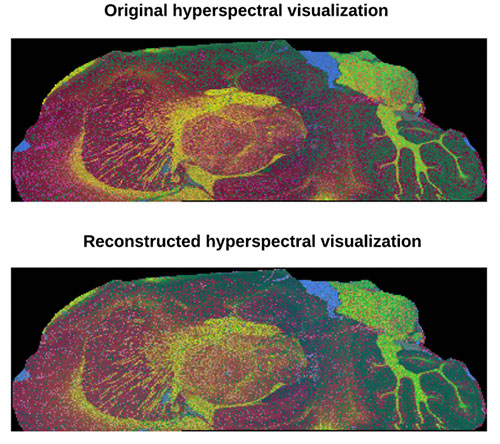 Top, hyperspectral visualization with data from a standard 9-hour experiment compared with hyperspectral visualization with data from a proposed 1-hour experiment.
Researchers at the Beckman Institute for Advanced Science and Technology developed a
subspace mass spectrometry imaging approach that accelerates the speed of data acquisition — without sacrificing the quality — by designing a model-based reconstruction strategy.
Top, hyperspectral visualization with data from a standard 9-hour experiment compared with hyperspectral visualization with data from a proposed 1-hour experiment.
Researchers at the Beckman Institute for Advanced Science and Technology developed a
subspace mass spectrometry imaging approach that accelerates the speed of data acquisition — without sacrificing the quality — by designing a model-based reconstruction strategy.
The technique, which was developed using animal models, could have important implications for many applications, including analytical chemistry and clinical studies, with results available at a fraction of the time. It also can detect a wide range of biomolecules — from small molecules such as neurotransmitters and amino acids to larger molecules such as lipids or peptides.
The paper “Accelerating Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry Imaging Using a Subspace Approach” was published in the Journal of the American Society of Mass Spectrometry.
“Fourier transform-ion cyclotron resonance is a really powerful instrument, providing the highest mass resolution,” said Yuxuan Richard Xie, a bioengineering graduate student at the University of Illinois Urbana-Champaign, who is first author on the paper. “But one disadvantage of FT-ICR is it's very slow. So essentially, if people want to achieve a certain mass resolution, they have to wait days to acquire data sets. Our computational approach speeds up this acquisition process, potentially from one day to maybe one to two hours — basically a tenfold increase in data acquisition speed.”
 Yuxuan Richard Xie, a bioengineering graduate student at the University of Illinois Urbana-Champaign, is first author on a paper about a new computational mass spectrometry imaging method.
“Our method is changing the way that we acquire the data,” Xie said. “Instead of acquiring mass spectra per pixel, the technique recognizes the redundancy in the high-dimensional imaging data and uses a low-dimensional subspace
model to exploit this redundancy to reconstruct multispectral images from only a subset of the data.”
Yuxuan Richard Xie, a bioengineering graduate student at the University of Illinois Urbana-Champaign, is first author on a paper about a new computational mass spectrometry imaging method.
“Our method is changing the way that we acquire the data,” Xie said. “Instead of acquiring mass spectra per pixel, the technique recognizes the redundancy in the high-dimensional imaging data and uses a low-dimensional subspace
model to exploit this redundancy to reconstruct multispectral images from only a subset of the data.”
Xie collaborated with Fan Lam, an assistant professor of bioengineering, and Jonathan V. Sweedler, the James R. Eiszner Family Endowed Chair in Chemistry and the director of the School of Chemical Sciences, who are co-principal investigators on the paper. Daniel Castro, a graduate student in molecular and integrative physiology, also contributed.
“We have been using subspace models in our MRI and MR spectroscopic imaging work for a long time,” Lam said. “It is really nice to see that it also has great potentials for a different biochemical imaging modality.”
“The ability to acquire enhanced chemical information and the locations of the chemicals in a complex sample such as a section of a brain becomes enabling for our neurochemical research,” Sweedler said.
The subspace imaging concept was pioneered by Zhi-Pei Liang, a professor of electrical and computer engineering and full-time Beckman faculty member, who is a world-leading expert in MRI and MRSI.
The research continues as researchers seek to apply the technique to 3D imaging. “(The approach) could have a much larger impact for the scientific community for 3D imaging of larger areas, such as the brain,” Xie said. “Because if we do 50 slices on FT-ICR, it would take weeks right now, but (with this technique) we can achieve decent coverage maybe within days.
“I believe that computational imaging, especially the data driven approach, is like a new shining star. It's getting more and more powerful, and we should definitely utilize some of those techniques for chemical analysis of tissue through mass spectrometry imaging.”
Editor’s note: The paper “Accelerating Fourier Transform-Ion Cyclotron Resonance Mass Spectrometry Imaging Using a Subspace Approach” is online at https://doi.org/10.1021/jasms.0c00276.
Beckman Institute for Advanced Science and Technology