Article
Nearly all life on Earth depends on photosynthesis, the conversion of light energy into chemical energy. Oxygen-producing plants and cyanobacteria perfected this process 2.7 billion years ago. But the first photosynthetic organisms were likely single-celled purple bacteria that began absorbing near-infrared light and converting it to sulfur or sulfates about 3.4 billion years ago.
Found in the bottom of lakes and ponds today, purple bacteria possess simpler photosynthetic organelles—specialized cellular subunits called chromatophores—than plants and algae. For that reason, Klaus Schulten targeted the chromatophore to study photosynthesis at the atomic level.
As a computational biophysicist, Schulten unites biologists’ experimental data with the physical laws that govern the behavior of matter. This combination allows him to simulate biomolecules, atom by atom, using supercomputers. The simulations reveal interactions between molecules that are impossible to observe in the laboratory, providing plausible explanations for how molecules carry out biological functions in nature.
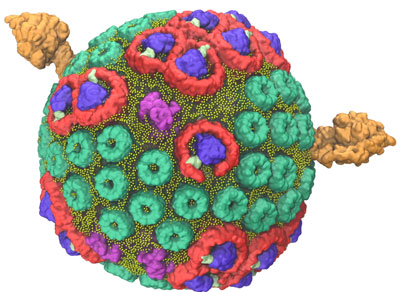
In 2014, a team led by Schulten used the Titan supercomputer, located at the US Department of Energy’s (DOE’s) Oak Ridge National Laboratory, to construct and simulate a single chromatophore. The soccer ball-shaped chromatophore contained more than 100 million atoms—a significantly larger biomolecular system than any previously modeled. The project’s scale required Titan, the flagship supercomputer at the Oak Ridge Leadership Computing Facility (OLCF), a DOE Office of Science User Facility, to calculate the interaction of millions of atoms in a feasible time frame that would allow for data analysis.
“For years, scientists have seen that cells are made of these machines, but they could only look at part of the machine. It’s like looking at a car engine and saying, ‘Oh, there’s an interesting cable, an interesting screw, an interesting cylinder.’ You look at the parts and describe them with love and care, but you don’t understand how the engine actually works that way,” Schulten said. “Titan gave us the fantastic level of computing we needed to see the whole picture. For the first time, we could go from looking at the cable, the screw, the cylinder to looking at the whole engine.”
Schulten’s chromatophore simulation is being used to understand the fundamental process of photosynthesis, basic research that could one day lead to better solar energy technology. Of particular interest: how hundreds of proteins work together to capture light energy at an estimated 90 percent efficiency.
Furthermore, the chromatophore project marks a shift in computational biophysics from analyzing the individual cell parts (e.g., a single protein) to analyzing the specialized systems of the cell (e.g., hundreds of proteins working together to carry out an autonomous function). This is a significant step toward the long-term goal of simulating an entire living organism.
Reconstructing Photosynthesis
When the purple bacteria Rhodobacter sphaeroides switches into photosynthetic mode, its inner membrane begins to change, bulging out into small, round vesicles that house the light-harvesting machinery.
Five major types of proteins arranged within two layers of lipids, or fats, contribute to the clockwork of processes that result in the conversion of light energy to adenosine triphosphate (ATP), the common fuel for cellular function across all branches of life.
During the initial steps of photosynthesis, two types of light-harvesting proteins absorb wavelengths of light that lift them into an excited state. This electronic excitation travels through the light-harvesting network to the third type of protein, known as the reaction center. Here, electrical energy is converted into an initial form of chemical energy. Molecules called quinols carry this chemical energy across the organelle to the fourth type of protein—the bc1 complex—where a charge separation process strips the quinol of electrons. This process triggers a current of protons in the fifth type of protein, known as ATP synthase, driving the molecule’s paddle wheel-like c-ring to produce ATP. (A detailed video of this process narrated by Schulten can be found here.)
Schulten’s team solved the 100-million-atom model under an allocation on Titan, awarded through the Innovative and Novel Computational Impact on Theory and Experiment, or INCITE, program.
The team used experimental data gathered from atomic force microscopy and the molecular dynamics code NAMD to build and calculate the forces exerted by the spherical chromatophore’s millions of atoms. The final chromatophore model measured 70 nanometers across and contained about 16,000 lipids and 101 proteins. To ensure the simulation mirrored nature, the chromatophore was submerged in a virtual 100 nanometer cube of water—equal to the largest particle size that can fit through a surgical mask—at room temperature and pressure.
Titan, a Cray XK7 with a peak performance of 27 petaflops (or 27 quadrillion calculations per second), proved pivotal in the early stages of the project. By offloading computationally demanding calculations to Titan’s GPUs, the team was able to achieve two to three times the performance of a CPU-only simulation. Additionally, NAMD scaled efficiently to more than 8,000 of Titan’s 18,688 nodes. The combination of robust hardware and effective software allowed the team to resolve its model in a few months, a task that would have taken more than a year on a smaller machine.
“Titan helped us immensely because you need to run the simulation for a certain amount of time and on a large number of processors in order to improve the model,” said team member Abhi Singharoy, a Beckman Institute Postdoctoral Fellow. “This project would have been impossible to do on another computer. Had we not achieved a stable model within the first few months, all our other aims would have been out of reach.”
Researchers earlier had leveraged Titan to resolve a flat, 20-million atom chromatophore patch as a stepping stone to the larger system. UIUC postdoctoral researchers Melih Sener and Danielle Chandler carried out the bulk of this initial work, arranging the proteins in accordance with atomic force microscopy data. A description of the 20-million-atom system was published in the August 2014 edition of Biophysical Journal.
Maximizing the Machine
With a stable model, the remarkable processes of the complete chromatophore—how its atoms move and coordinate within an active, energy-conversion system—can be studied in unprecedented detail.
Under a 150-million processor-hour allocation awarded through the 2015 INCITE program, the team is continuing to run its simulation and analyze the chromatophore for notable properties, such as the organelle’s optimum pH and salt concentration levels. Analysis and visualization of the organelle are being conducted using an application called VMD. Both VMD and NAMD originated with and continue to be developed by Schulten’s Theoretical and Computational Biophysics Group. The applications support a global community of users.
NAMD calculates the motion of the chromatophore’s atoms in time steps of 2 femtoseconds, or 2,000 trillionths of a second. At this timescale, a 1-day, 4,000-processor run on Titan nets 16 nanoseconds (16 million femtoseconds), or 16 billionths of a second, of simulation time. By continuing to run the simulation—with the goal of capturing a microsecond (1,000 nanoseconds) of simulation time—slower-moving processes of the biomolecular system can be observed.
“With the time we’ve been allotted on Titan, we will actually be able to see a quinol move and the charge being transferred,” Singharoy said.
The OLCF is helping Schulten’s team manage and analyze the terabytes of data produced by the project. Typically, simulation data are transferred to a project’s home institution for analysis. Because of the size of the chromatophore, a transfer job can take days to complete—even using world-class networking resources.
To overcome this bottleneck, the OLCF worked to enable the graphics capabilities of Titan’s GPUs. Opening up Titan for hardware-accelerated graphics makes it possible for users to run VMD’s graphical interfaces in parallel with NAMD from any location, creating a near real-time “window” into calculations being performed on Titan. Such remote visualization would make it easier for researchers to view and manipulate their data while eliminating the file transfer. This VMD capability is expected to be available to users soon.
“We’re making the scientists much more productive,” said John Stone, a senior research programmer at UIUC and VMD’s lead developer. “Essentially, it will be like having a petascale computer on your laptop.”
Beckman Institute for Advanced Science and Technology