Interphase Chemical Mapping of Carbon Fiber-Epoxy Composites by AFM-IR Spectroscopy
Chris Montgomery, Autonomous Materials Systems Group
Abstract:
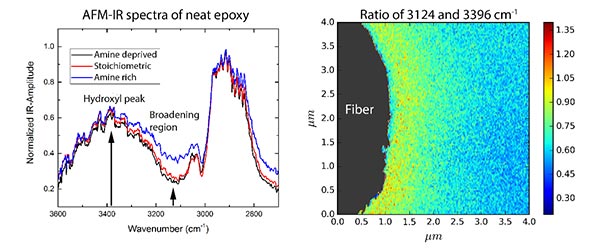
The properties and performance of carbon fiber reinforced polymer matrix composites are highly influenced by the chemical interactions of the fiber/matrix interface region. Many researchers have hypothesized that the presence of carbon fibers in epoxy causes a chemical gradient to form around the fiber during the curing stage. However, the chemical makeup of the interphase region and how it influences composite behavior is yet to be fully understood. One of the obstacles has been the lack of experimental techniques available to chemically probe the interphase with sufficient resolution.
High-resolution spectroscopy techniques recently developed to study biological processes at the cellular level also have significant potential for characterization of synthetic materials systems. Here, we apply atomic force microscopy based infrared (AFM-IR) spectroscopy to investigate the interphase region developed between carbon fibers and epoxy matrix in an aerospace-grade composite material. The working principle of the technique will be reviewed and key findings will be highlighted. AFM-IR spectra of the bulk matrix material are in excellent agreement with spectra collected by traditional FTIR in both the low wavenumber range (900-1700/cm) and in the high wavenumber range (> 2500/cm). Chemical maps of the interphase region provide preliminary evidence for distinct variations in matrix chemistry near the fiber surface in comparison to bulk.
Genome-Scale Metabolic Models for Your Gut
ShengShee (Seth) Thor, a doctoral student in biophysics
Abstract:
The human gut microbiota is a complex microbial community that biomedical researchers have yet to fully understand. Much of the research has been dedicated to studying the activities of gut bacteria exclusively, leaving eukaryotic and archaeal gut species activity largely uncharacterized. Growing evidence continues to suggest intimate connections between imbalanced microbiota dynamics and numerous human ailments. However, these connections remain unclear without the aid of good computational models for probing microbiota behavior across all three domains of life. In this talk, I will first provide an overview of the current human gut microbiota research under two different lenses. Afterwards, I will discuss my research in developing genome-scale metabolic models specifically for relevant methanogenic archaea (methanogens) under the context of human gut microbiota research.
Epigenetics with Semiconductor Nanotechnology
Aditya Sarathy, Nanolectronics and Nanomaterials Group
Abstract:
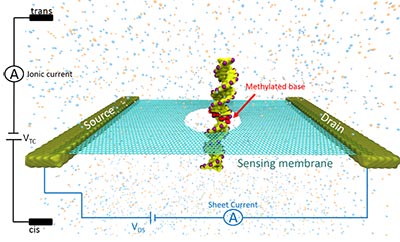
DNA methylation is an epigenetic modification involving the addition of a methyl group to DNA, which is heavily involved in gene expression and regulation, thereby critical to the progression of diseases such as cancer. In this work we show that detection and localization of DNA methylation can be achieved with nanopore sensors made of two-dimensional (2D) materials such as graphene and Molybdenum di-Sulphide . We label each DNA methylation site with a methyl-CpG binding domain protein (MBD1), and combine molecular dynamics simulations with electronic transport calculations to investigate the translocation of the methylated DNA-MBD1 complex through 2D material nanopores under external voltage biases. The passage of the MBD1-labeled methylation site through the pore is identified by dips in the current blockage induced by the DNA strand, as well as by peaks in the transverse electronic sheet current across the 2D layer. The position of the methylation sites can be clearly recognized by the relative positions of the dips in the recorded ionic current blockade and large deviations in the transverse sheet conductances. We define the spatial resolution of the 2D material nanopore device as the minimal distance between two methylation sites identified within a single measurement, which is 15 base pairs by ionic current recognition, but as low as 10 base pairs by transverse electronic conductance detection, indicating better resolution with this latter technique. The present approach opens a new route for precise and efficient profiling of DNA methylation.